A. RNA-protein recognition
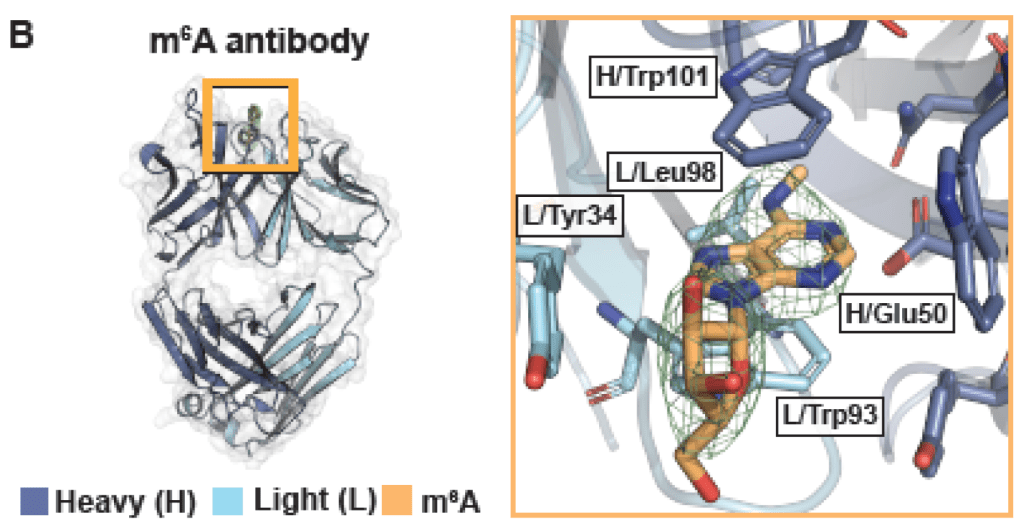
We’re interested in how proteins recognize RNA, with a particular focus on RNA modifications. Can we develop new methods to illuminate the functions of understudied RNA modifications? What are their biochemical and biological functions in mRNA? The work has been and continues to be highly collaborative. (Funding: NIH, NSF)
key and current publications
- Angelo M, Zhang W, Vilseck JZ, Aoki ST. In silico λ-dynamics predicts protein binding specificities to modified RNAs. Nucleic Acids Res. 2025 Feb 27;53(5):gkaf166. DOI: 10.1093/nar/gkaf166. PDF
- Angelo M, Bhargava Y, Kierzek E, Kierzek R, Hayes RL, Zhang W, Vilseck JZ, Aoki ST. Accurate in silico predictions of modified RNA interactions to a prototypical RNA-binding protein with λ-dynamics. RNA. 2025 Sep 16;31(10):1460-1471. DOI: 10.1261/rna.080367.124. PDF
B. RNA-protein complex assemblies
and their biochemical functions

RNA-protein complexes can assemble into higher-ordered structures, sometimes forming biomolecular condensates. How do these condensates assemble and what is their structural order? What are the biochemical functions of these condensates? This investigation uses biochemistry and structural biology methods. (Funding: NIH)
key and current publications
- Sandoval CH, Borchers C, Yang B, Boyd B, Hundley HA, Aoki ST. eIF4E assembly into C. elegans germ granules is essential for its repressive function. bioRxiv [Preprint]. 2025 Sep 25:2025.09.24.678427. DOI: 10.1101/2025.09.24.678427. PDF
- Aoki ST, Crittenden S, Lynch T, Bingman CA, Wickens M, Kimble J.C. elegans germ granules require both assembly and localized regulators for mRNA repression. Nat Commun. 2021 Feb 12;12(1):996. DOI: 10.1038/s41467-021-21278-1. PDF
C. Biological functions
of RNA-protein complexes
in situ
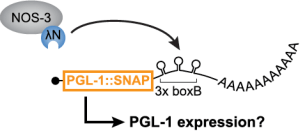
We’re interested in how RNA-protein assemblies function in situ, or their authentic tissue. How does RNA-protein complex assembly connect to its function in germ cells destined to become future progeny? The C. elegans germline continues to be our muse. (Funding: NIH)
key and current publications
- Borchers C, Osburn K, Roh HC, Aoki ST. In vivo pulse-chase in Caenorhabditis elegans reveals intestinal histone turnover changes upon starvation. J Biol Chem. 2025 Jul;301(7):110299. DOI: 10.1016/j.jbc.2025.110299. PDF
- Doenier J, Lynch TR, Kimble J, Aoki ST. An improved in vivo tethering assay with single molecule FISH reveals that a nematode Nanos enhances reporter expression and mRNA stability. RNA. 2021 Jun;27(6):643-652. PDF
D. Identify and develop hidden talent

An educational interest is discovering new strategies to recognize and nurture science talent in our community. How can we find science talent in our community? How can we effectively develop trainees for STEM careers? (Funding: NSF)
key and current publications
- Miller G, Aoki S. A Primer for Junior Trainees: The Structure, Binding, and Therapeutic Applications of Immunoglobulin G. Preprints: Preprints; 2025. PDF
- Herrera-Sandoval C, Borchers C, Aoki ST. An effective Caenorhabditis elegans CRISPR training module for high school and undergraduate summer research experiences in molecular biology. Biochem Mol Biol Educ. 2024 Jul 27. doi: 10.1002/bmb.21856. PDF
- Aoki ST. ‘The five love languages’ in science mentoring. ASBMB Today, 1/17/2020. https://www.asbmb.org/asbmb-today/opinions/011720/the-five-love-languages-in-science-mentoring.
E. Funding


